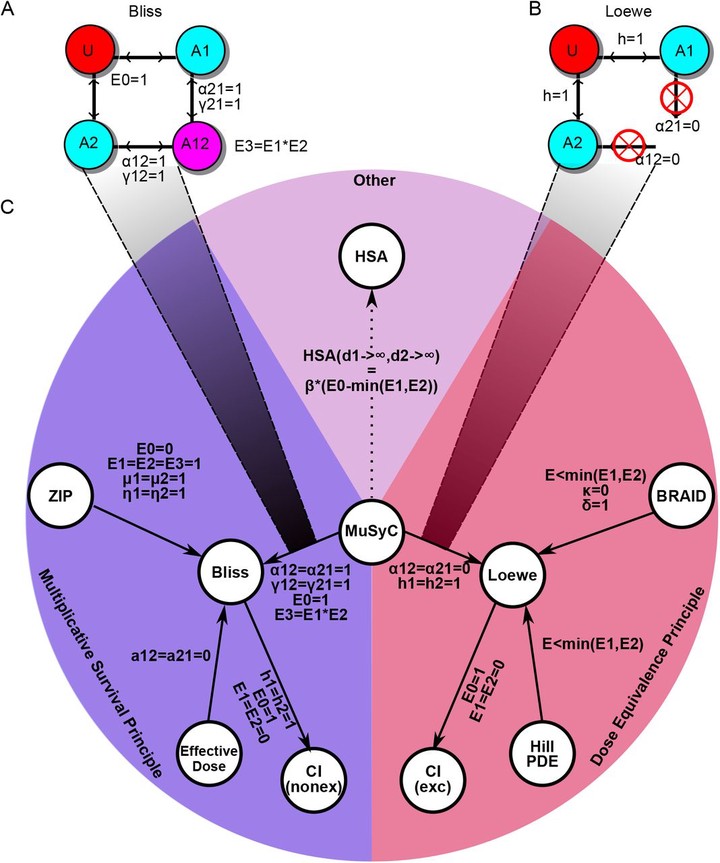
Abstract
Drug combination discovery depends on reliable synergy metrics; however, no consensus exists on the appropriate synergy model to prioritize lead candidates. The fragmented state of the field confounds analysis, reproducibility, and clinical translation of combinations. Here we present a mass-action based formalism to accurately measure the synergy of drug combinations. In this work, we clarify the relationship between the dominant drug synergy principles and show how biases emerge due to intrinsic assumptions which hinder their broad applicability. We further present a mapping of commonly used frameworks onto a unified synergy landscape, which identifies fundamental issues impacting the interpretation of synergy in discovery efforts. Specifically, we infer how traditional metrics mask consequential synergistic interactions, and contain biases dependent on the Hill-slope and maximal effect of single-drugs. We show how these biases systematically impact the classification of synergy in large combination screens misleading discovery efforts. The proposed approach has potential to accelerate the translatability and reproducibility of drug-synergy studies, by bridging the gap between the curative potential of drug mixtures and the complexity in their study.